个人资料
教育经历2006.08-2010.05 美国佛罗里达大学化学系, 物理化学方向, 主攻大分子的计算机模拟, 获博士学位, 导师:Professor Kenneth M. Merz (Editor-in-chief of the Journal of Chemical Information and Modeling (JCIM,IF=4.304) since 2014. Now: Director of iCER of Michigan State University ) 2003.09-2006.06 南京大学化学系, 理论与计算化学方向, 获硕士学位,导师: 张增辉教授 1999.09-2003.06 南京大学理科强化班, 物理方向, 获学士学位 1996.09-1999.06 江苏省苏州中学,高中 1993.09-1996.06 江苏省苏州中学,初中 工作经历2021-至今 上海市分子智造前沿科学研究基地,副主任(2021-2024),主任(2024-至今),上海分子治疗与新药创制工程技术研究中心副主任(2024-至今),LSZN,SMFZ,CZS(2024-至今) 2015.12-至今 教授(破格),博导(2017.06-),二级教授(2023-),化学与分子工程学院,华东师范大学 2013.09-至今 兼职教授,上海纽约大学计算化学研究中心 2011.07-2015.12 副教授,精密光谱科学与技术国家重点实验室,华东师范大学 2014.07-2014.10;2015.08-2015.10 访问学者,美国明尼苏达大学化学系,合作教授:Professor Donald Truhlar(美国科学院院士) 2011.10-2012.09 博士后研究员,美国伊利诺伊大学香槟分校(UIUC)化学系,导师:Professor So Hirata (国际量子分子科学院院士) 2011.03-2011.07 专职研究员,华东师范大学物理系 个人简介2003年与2006年分别于南京大学获得本科和硕士学位,2010年5月毕业于美国佛罗里达大学化学系,获得理学博士学位。2011年至2012年在美国伊利诺伊大学香槟分校化学系担任博士后研究员。在硕士研究生阶段师从张增辉教授,从事理论与计算化学的研究,发展了全量子计算蛋白质总能量的分块方法,在博士研究生阶段师从Kenneth Merz教授,主要研究方向是大分子体系的Divide-and-conquer的从头算量子化学方法的发展,以及基于量子力学方法的蛋白质NMR化学位移计算和量子化学软件的开发。在博士后阶段师从So Hirata教授,主要课题是将分块的量子化学方法应用于固态,聚合物和材料,以及发展有限温度下的电子结构理论,一共编写过三个量子化学计算软件:QUICK,AF-NMR和EDISON2.0。2011年8月进入华东师范大学,获得副教授职称,2015年12月破格提升为教授。在化学与分子工程学院从事理论与计算化学和生物学的研究,主要研究方向为复杂大分子体系的线性标度量子化学方法,生物大分子NMR参数的精确量子化学计算方法,密度泛函方法和深度机器学习方法的发展。2014,2015年在美国明尼苏达大学任访问学者,与美国科学院院士Donald Truhlar教授合作发展新的密度泛函方法。目前已在Nat. Comput. Sci.(2篇),PNAS(IF=11.21,3篇),Sci. Adv.(IF=14.98,1篇),Nat. Commun.(IF=17.69,6篇),JACS(IF=16.38,8篇),Angew. Chem. Int. Ed. (IF=16.82,10篇),Energy Environ. Sci.(IF=32.4,1篇),Matter(IF=17.3,1篇),Acc. Chem. Res.(IF=22.38,2篇),WIRES Comput. Mol. Sci.(IF=11.5,1篇),Cell Research(IF=46.30,1篇),Advanced Materials(IF=27.4,1篇),Journal of Clinical Investigation(IF=13.3,1篇),Chem. Sci.(IF=9.97,2篇),ACS Catalysis(IF=13.1,1篇),Small(IF=15.153,3篇),ACS Appl. Mater. Interfaces(IF=10.38,1篇),IEEE Computational Intelligence Magazine(IF=9,1篇),ACS Sustainable Chem. Eng.(IF=9.22,1篇),J. Membr. Sci.(IF=8.74,1篇),J. Med. Chem.(IF=8.04,1篇),Analytical Chemistry(IF=7.4,1篇),J. Phys. Chem. Lett.(IF=6.89,3篇),NPJ Quantum Materials(IF=7.03,1篇),Food Chemistry (IF=7.51,1篇),Antiviral Research(IF=7.6,1篇),Chem. Comm.(IF=6.07,2篇),ACS Applied Nano Materials(1篇),Nanoscale (1篇),Nanoscale Advances (1篇),J. Chem. Theory Comput.(IF=6.01,17篇),J. Chem. Inf. Model.(IF=6.16,9篇),ChemCatChem(IF=5.50,1篇),Inorganic Chemistry(IF=4.3,1篇),Macromolecules(IF=4.40,1篇),J. Phys. Chem. C(IF=4.22,2篇),Phys. Chem. Chem. Phys.(IF=3.57,12篇),Cryst. Growth Des. (IF=4.01,1篇),Phys. Rev. Mater.(IF=3.99,1篇),J. Chem. Phys.(IF=3.49,12篇),J. Phys. Chem. B(IF=2.99,7篇),J. Phys. Chem. A(IF=2.78,11篇),J. Comput. Chem.(IF=3.38,1篇),Journal of Organic Chemistry(IF=3.6,1篇),J. Comput.-Aided Mol. Des.(IF=3,1篇)等国际著名杂志上发表SCI论文195篇,其中第一作者和通讯作者身份署名文章143篇(第一作者12篇,通讯作者130篇),参编国际专著4部,2篇文章入选全球ESI 1%高被引论文。文章总的被引用数超过8000次,h-index达到45,授权专利8项,软件著作权4项。2024年入围高性能计算应用“戈登贝尔奖”,2024年荣获CCF HPC China2024超算年度最佳应用奖,2023年获得教育部“长江学者”特聘教授,2019年获得“国家优秀青年基金”资助(结题优秀)与中国化学会“唐敖庆理论化学青年奖”,并入选上海市青年拔尖人才,上海市普陀区青年英才和华东师范大学紫江优秀青年学者等人才计划支持。获得美国化学会优秀导师奖,中国化学会“中国青年化学家元素周期表”氪元素代言人,华东师范大学研究生教育卓越育人奖(优秀导师奖)。现任中国化学会理论化学专业委员会委员,中国化学会计算(机)化学专业委员会委员,中国生物信息学会生物信息与药物发现专业委员会委员,上海市药学会第一届人工智能药学专业委员会委员,科技部科技评估中心重点领域咨询专家,Communications in Computational Chemistry期刊副主编,J. Chem. Inf. Model.杂志的顾问编委。指导的学生获上海市“超级博士后”,博士后国资计划,国自然青年基金,博士后面上项目,海外博士后引进计划,“上海市大学生化学化工优秀论文一等奖”,“美国化学会会议优秀报告奖”和国家奖学金等。现为华东师范大学二级教授,任上海市分子智造前沿科学研究基地主任,上海分子治疗与新药创制工程技术研究中心副主任,上海市未来学科(量子科学与技术)课题负责人,于2023年提出并发布适用于化学领域和分子逆合成的化学大语言模型ChemGPT 1.0,并于2024年发布ChemGPT 2.0的升级版本(https://chemgpt.lrcwtech.com/)。受邀在Nat. Comput. Sci.上发表观点文章。
社会兼职2025- 烟草行业卷烟烟气重点实验室学术委员会委员 2024- 科技部科技评估中心重点领域咨询专家 2024- 上海市药学会第一届人工智能药学专业委员会委员 2024- Associate Editor, Communications in Computational Chemistry, https://global-sci.com/journal/28/communications-in-computational-chemistry 2024- 中国化学会高级会员 link: 何晓 - 高级会员 - 中国化学会 (chemsoc.org.cn) 2022- 美国化学会ACS旗下Journal of Chemical Information and Modeling期刊(2020年影响因子4.956)的Editorial Advisory Board Member 中国化学会理论化学专业委员会委员 中国生物信息学会生物信息与药物发现专业委员会委员 Science Advances, Nature Communications, Nature Computational Science, National Science Review, ACS Catalysis, Journal of Materials Chemistry A, Sustainable Chemistry and Pharmacy, Nano Letters, Precision Chemistry, Journal of Agricultural and Food Chemistry, Journal of Materials Chemistry B, npj Computational Materials, ACS Applied Nano Materials, Sensors and Actuators B: Chemical, Chemistry-An Asian Journal, Food Chemistry, Nanoscale, Energy Advances, Sustainable Chemistry and Pharmacy,Frontiers in Pharmacology, Chinese Science Bulletin, Membranes, Advanced Sustainable Systems, Chemical Journal of Chinese Universities, Journal of Physics: Condensed Matter, Informatics in Medicine Unlocked, Computational and Theoretical Chemistry, Chemical Society Reviews, Journal of the American Chemical Society, Chemical Science, The Journal of Physical Chemistry Letters, Frontiers in Molecular Biosciences, Crystal Growth & Design, ChemistrySelect, Chinese Journal of Chemistry, Journal of the Chinese Chemical Society, Frontiers in Chemistry, New Journal of Chemistry, New Journal of Physics, Materials Today Bio, Expert Review of Proteomics, Computational and Structural Biotechnology Journal, Communications Chemistry, Photochemistry and Photobiology, Biophysical Journal, ACS Omega, Chemical Communications, Analyst, Scientific Reports, Journal of Computational Chemistry, Journal of Theoretical Biology, Biophysical Chemistry, Chemical Data Collection, Journal of Chemical Physics, Chemistry-A European Journal, International Journal of Molecular Sciences, Proteins, PeerJ, RSC Advances, Molecular Simulation, Journal of Molecular Modeling, Journal of Molecular Graphics and Modelling, Molecular BioSystems, Journal of Chemical Information and Modeling, Journal of Chemical Theory and Computation, Journal of Computer-Aided Molecular Design, Journal of Physical Chemistry A/B, Physical Chemistry Chemical Physics, Organic Letters, Journal of Theoretical and Computational Chemistry, Chemical Physics Letters 等杂志审稿人 研究方向
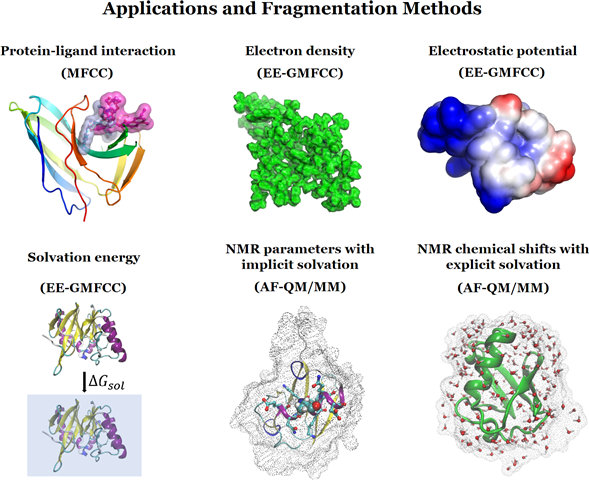
开授课程2012.10-2013.01; 2013.09-2014.01; 2014.10-2015.01; 2015.10-2016.01 华东师范大学研究生课程-分子体系的量子力学方法和应用 2016.09-2016.12; 2017.09-2017.12; 2018.09-2018.12; 2019.09-2019.12; 2020.09-2020.12; 2021.09-2021.12; 2022.09-2022.12; 2023.09-2023.12 Foundations of Chemistry I Recitation 2022.02-2022.05 Foundations of Chemistry II Recitation 2018.2-2018.5; 2019.2-2019.5;2025.2-2025.5 Foundations of Chemistry II 2019.09-2019.12; 2020.09-2020.12 General Physics I 2021.01-2021.05 Foundations of Physics Honors II 2017.02-2017.07; 2018.02-2018.07; 2019.02-2019.07;2020.02-2020.07; 2021.02-2021.07; 2022.02-2022.07; 2023.02-2023.07;2024.02-2024.07; 02/2025-07/2025; 02/2026-07/2026;02/2027-07/2027;02/2028-07/2028;02/2029-07/2029;02/2030-07/2030; 研究生课程-计算化学 2019.09-2020.01; 2020.09-2021.01; 2021.09-2022.01; 2022.09-2023.01; 2023.02-2023.07;2024.02-2024.07; 02/2025-07/2025; 02/2026-07/2026;02/2027-07/2027;02/2028-07/2028;02/2029-07/2029;02/2030-07/2030 本科生课程-分子计算机模拟与应用(Starting from 2025, 智能分子模拟与计算化学前沿应用) 09/2024-01/2025; 09/2025-01/2026; 09/2026-01/2027;09/2027-01/2028;09/2028-01/2029;09/2029-01/2030 研究生课程-药物智造 科研项目2013.01~2014.12 Startup fund for junior faculty of State Key laboratory of Precision Spectroscopy, PI 2014.01~2016.12 National Natural Science Foundation of China, PI 2013.01~2014.12 The Fundamental Research Funds for the Central Universities of ECNU, PI 2014.01~2016.12 Specialized Research Fund for the Doctoral Program of Higher Education, PI 2015.09~2016.09 Ab initio calculation on biological systems, PI 2017.01~2020.12 National Natural Science Foundation of China, PI 2016.07~2021.06 Ministry of Science and Technology of China, Co-PI 2017.01~2018.12 Shanghai Young Top-notch Talent Program, PI 2017.01~2018.12 Young Talent in Putuo District, PI 2017.03~2019.03 JRI Seed Grants for Research Collaboration from NYU-ECNU Joint Research Institutes at NYU Shanghai, PI 2018.01~2020.12 National Natural Science Foundation of China, PI 2018.07~2021.06 Natural Science Foundation of Shanghai, PI 2019.06~2022.07 US NSF OISE (Office of International Science and Engineering) program, co-PI 2020.01~2022.12 NSFC Excellent Young Scholar, PI 2019.07~2019.12 The Fundamental Research Funds for the Central Universities of ECNU, PI 2019.08~2025.07 Start-up fund from ECNU, PI 2019.10~2020.12 Fund from local District 1/2, PI/Co-PI 2020.01~2024.12 Ministry of Science and Technology of China, Co-PI 2020.06-2023.12 Fund from Shanghai Engineering Research Center of Molecular Therapeutics and New Drug Development, sponsered by the Fundamental Research Funds for the Central Universities, Tier 1/2 2020.07-2020.12 Fund from School of Chemistry and Molecular Engineering, Tier 1 2021.01-2025.12 Fund from Research Center of Shanghai(8.0+7.5+1.1) 2022.07-2025.07 Fund from University, tier 1/2 2022.01-2022.12-2023.12 Chemistry department*1(5), University*2(2+10*2) 2023.01~2026.12 National Natural Science Foundation of China, PI 2022.01~2024.12 Shanghai Engineering Research Center of Molecular Therapeutics and New Drug Development (15) 2023.04~2026.03 Natural Science Foundation of Shanghai, PI 2023 Minister of Education-ECNU (10) Co-PI 2023-2025 Chongqing (10) 2023-2026 Syngenta (30) 2023 Saike (30) 2024 Bojie(45) 2024 ***(2.3) 2024 University(4.0),(1.5,0.3,0.6:Rui) 2024 JiDi 1/y ...... See Xiao's PC for more info. 学术成果PUBLICATIONS (*corresponding author, #co-first author) (Total citations > 8000, h-index: 45) Google Scholar: http://scholar.google.com.hk/citations?user=L37jIigAAAAJ&hl=en 206) J. Cao, Z.A. Chen, H.C. Li, C. Liu, Y.T. He, H.B. Zhang, L.N. Xu*, H.P. Xiao, X. He* and G.Y. Fang*, Intelligent design and simulation of high-entropy alloys via machine learning and multi-objective optimization algorithms. J. Chem. Theory Comput. In press (2025) 205) J.H. Zhang#, Z. Wan#, Q.Y. Gu, J.B. Zhu, J. Zhong*, X. He*, J.R. Yang*, π-π Interactions Dictate the Growth of Aromatic Organic Aerosol. J. Phys. Chem. A In press (2025) 204) S.Q. Li#, J Tuo#, Z. Wan#, S. Zhang, M.Y. Yu, Y. Ma, R.S. Peng*, H. Xu, J.G. Jiang, S.F. Chen, X. He*, P. Wu*, Minimizing Sinusoidal Channels of ZSM-5 for Propene Production from Molecular Traffic Controlled Cracking Reaction. Small 2503660 (2025) 203) D.Y. Hou#, S. Zhang#, M.Y, Ma, H.B. Lin, Z. Wan, H. Zhao, R.A. Zhou, X. He*. X. Wei*, D.W. Ju* and X. Zeng*, A Hyperbolic Discrete Diffusion 3D RNA Inverse Folding Model for functional RNA design. J. Chem. Inf. Model. In press (2025) 202) Y. Zhang,H. Hao*, X. He, A.M. Zhou, RetroEA: An efficient evolutionary algorithm for retrosynthetic route planning. Swarm and Evolutionary Computation (IF=8.2) 96, 101967 (2025) 201) J.F. Liu*,X.C. Zhou, X. He*, Structure making and breaking effects of ions on the anomalous diffusion of water revealed by machine learning potentials. Phys. Chem. Chem. Phys. In press (2025) 200) X.X. Shen#,Z. Wan#,L.F. Wen,L.C. Sun,J. Yang,X. Tang,Shing-Ho J. Lin,X. He,M.S. Chen,X. Wei*, PAIRWISE DISTANCE DISTRIBUTION GRAPH TRANSFORMERS FOR CRYSTAL MATERIAL PROPERTY PREDICTION. IJCAI In press (2025) 199) J.F. Li#, W.Q. Tang#, J.B. Zhu, J.R. Yang* and X. He*, Hydroxymethanesulfonate formation accelerated at the air-water interface by synergistic enthalpy-entropy effects. Nat. Commun. 16, 5187 (2025) 198) Z.C Liu, J.J. Li, C. Zhao, Z.H. Zhang, P.C. Wu, J.Q. Chen, X. He, S.J. Zhang,* and Yang Tian*, Molecular Engineering Enables Bright Carbon Dots for Super-Resolution Fluorescence Imaging and In Vivo Optogenetics. Advanced Materials 37, 2410786 (2025) 197) S.W. Chen, J.B. Zhu, J.F. Li, P. Guo*, J.R. Yang* and X. He*, Entropy-driven difference in interfacialwater reactivity between slab and nanodroplet. Nat. Commun. 16, 5250 (2025) 196) G.Y. Wu, L. Hu, L. Zhu, Z.Y. Xu, W.L. Hu, L.R. Hu*, Y. Qin, X. He, B.Z. Tang* and Z. Lu*, FRET-mediated Photoswitchable Catalytic Metallacage with Tunable Activity for Organic Dye Degradation. Inorganic Chemistry 64, 6865 (2025) 195) H. Wang, H.W. Zhang, J.H. Zhai, Y.Y. Liu*, B. Song, X. He, Y.F. Ge and D. Li*, Breaking Hydrogen Bond Network Enhances Oxygenic Photosynthesis of Photosystem II. Fundamental Research In press (2025) 194) Y.T. Li, D.Y. Xiong, J.B. Zhu, Y.L. Mou, J.R. Yang* and X. He*, Dynamic Mechanism of Short Peptide Additive Regulating Solvation Microenvironment of Zinc Ions. Batteries & Supercaps e202400735 (2025) 193) D.Y. Xiong#, Y.F Ming#, Y.T. Li, S.H. Li, K.X. Chen, J.F. Liu*, L.L. Duan*, H.L. Li, M. Li* and X. He*, EvoNB: A Protein Language Model-Based Workflow for Nanobody Mutation Prediction and Optimization. Journal of Pharmaceutical Analysis 15, 101260 (2025) 192) S.W. Chen, Z.H. Zhang, Y.T. Li, Y.J. Chen, J.R. Yang*, X. He* and L. Zhang*, Implications of weaving pattern on the material properties of two-dimensional molecularly woven fabrics. Matter 8, 102050 (2025) 191) Z.A. Chen, Z.J. Meng, T. He, H.C. Li, J. Cao, L.N. Xu*, H.P. Xiao, Y.Y. Zhang, X. He* and G.Y. Fang*, Crystal structure prediction meets artificial intelligence. J. Phys. Chem. Lett. 16, 2581 (2025) 190) D.H. Dai, Y.P. Liu*, X. He* and J.F. Mao*, Dynamic Nuclear Polarization and Chemically Induced Hyperpolarization: Progress, Mechanisms, and Opportunities. Magnetic Resonance Letters In press (2025) 189) X.J. Zhou*, L. Li, J. Xue, F. Wang and X. He*, Theoretical Study on Kinetics of Secondary Oxygen Addition Reactions for N-butyl Radicals. J. Phys. Chem. A 129, 1441 (2025) 188) X.W. Wang, D.Y. Xiong, Y.Q. Zhang, J.H. Zhai, Y.C. Gu and X. He*, The Evolution of the Amber Additive Protein Force Field: History, Current Status, and Future. J. Chem. Phys. 162, 030901 (2025) 187) Y.L. Mou, Y.Z. Jiang, X. He*, L.J. Zhang* and J.R. Yang*, Dynamic Modulation of Ions Solvation Sheath by Butyramide as Electrolyte Additive in Aqueous Batteries. J. Phys. Chem. B 129, 423 (2025) 186) X.H. Wang, Z.Y. Zhou, X. He*, Z.R. Liu and Z.X. Sun*, True Dynamics of Pillararene Host-Guest Binding. J. Chem. Theory Comput. 21, 241 (2025) (Virtual Special Issue on “Developments of Theoretical and Computational Chemistry Methods in Asia”) 185) Y.S. Liu#, J.F. Liu#, W. Chen, Y.C. Li, X. He* and Q. Yang*,Structure-based virtual screening of novel chitin synthase inhibitors for the control of Phytophthora sojae. Pest Management Science 81, 777 (2025) 184) Z. Zhou#, Y. Feng#, M. Jiang, Z. Yao, J. Wang, F. Pan, R. Feng, C. Zhao, Y. Ma, J. Zhou. L. Sun, X. Sun, C. Zhan, X. He*, K. Jiang*, J.H. Yu* and Z.Q. Yan*, Ionizable polymeric micelles (IPMs) for efficient siRNA delivery. Nat. Commun. 16, 360 (2025) 183) J.N. Liu#, Y.Y. He#, Y.Q. Li#, X.X. Chen, H.T. Li, J.R. Xu, H.Z. Chen, Y. Wang, X. He*, S.Y. Liu* and L.L. Chen*, Exploring Bifunctional Molecules: Anti-SARS-CoV-2 and Anti-Inflammatory Activities via Structure-Based Virtual Screening and Biological Evaluation, International Journal of Biological Macromolecules 287, 138529 (2025) 182) X. Wang#, C. Zhao#, M.Y. Yang#, J.H. Baek, Z. Meng, B. Sun, A.H. Yuan, J.B. Baek*, X. He*, Y. Jiang* and M.F. Zhu*, Bioinspired Photothermal Metal-Organic Framework Cocrystal with Ultra-fast Water Transporting Channels for Solar-driven Water Evaporation. Small 21, 2407665 (2025) 181) E. Huang, S. Liu, X. Zhou, L. Zhong, S. Ma, R. Wang, H. Li, J.R. Yang, X. He, W.R. Dong* and X.M. Yang, Vibrational Spectroscopy and Structural Characterization of Neutral and Cationic Hydroxyacetone. J. Mol. Struct. 1321, 139871 (2025) 180) H.H. Shang*, Y. Liu*, Z.K. Wu, Z.C. Chen, J.F. Liu, M.Y. Shao, Y.Z. Li, B.W. Kan, H.M. Cui, X.B. Feng, Y.Q. Zhang, D.G. Truhlar, H. An, X. He* and J.L. Yang*, Pushing the Limit of Quantum Mechanical Simulation to the Raman Spectra of a Biological System with 100 Million Atoms, SC24, November 17-22, 2024, Atlanta, Georgia, USA (2024) (Finalist for the 2024 Gordon Bell Prize) 179) Q.N. Liu#, J.N. Huang#, H. Ding#, Y. Tao#, J.L. Nan, C.C. Xiao, Y.C. Wang, R.R. Wu, C. Ni, Z.W. Zhong, W. Zhu, J.H. Chen, C.Y. Zhang, X. He, D.Y. Xiong, X.Y. Hu* and Jian’an Wang*, Flavin-containing monooxygenase 2 confers cardioprotection in ischemia models through its disulfide-bond catalytic activity. Journal of Clinical Investigation 134, e177077 (2024) 178) Z.A. Chen, H.C. Li, C. Zhang, H.B. Zhang, Y.X. Zhao, J. Cao, T. He, L.N. Xu*, H.P. Xiao, Y. Li, H.Z. Shao, X.Y. Yang, X. He* and G.Y. Fang*, Crystal Structure Prediction Using Generative Adversarial Network with Data-Driven Latent Space Fusion Strategy. J. Chem. Theory Comput. 20, 9627 (2024) 177) Y.L. Chen, Z. Wan*, Y.Y. Li, X. He*, X. Wei and J. Han*, Graph Curvature Flow-based Masked Attention. J. Chem. Inf. Model. 64, 8153 (2024) 176)Z. Wan#, Z.Y. Chen#, H. Chen, Y.Z. Jiang, J.H. Zhang, Y.D. Wang, J.D. Wang, H. Sun, H. Sun, J.H. Zhu, L.Y. Yang, W. Ye, S.K. Zhang, X. Xie, Y. Zhang, X.D. Zhuang,* X. He* and J.R. Yang*, XRDMatch: A Semi-Supervised Learning Framework to Efficiently Discover Room Temperature Lithium Superionic Conductors. Energy Environ. Sci. (Impact Factor=32.4) 17, 9487 (2024) 175) Y.L. Li, Y.H. Chen and X. He*, Teaching Spin Symmetry while Learning Neural Network Wavefunctions. Nat. Comput. Sci. 4, 884 (2024) 174) X.L. Qiao, S.J. Zhai, J.W. Xu, H.B. He, X. He, L.R. Hu* and S.H. Gao*, Asymmetric Photo-induced Excited-state Nazarov Reaction. J. Am. Chem. Soc. 146, 29150 (2024) 173) Y.Z. Jiang, D.Y. Xiong, Z. Wan, J.R. Yang* and X. He*, Mechanism-guided Rational Design of Anode Coatings for Aqueous Zinc Ion Batteries. ChemPhysChem 25, e202400231 (2024) 172) Y.Y. Shu, J.D. Yue, Y. Li, Y. Yin, J.X. Wang, T. Li, X. He, S.P. Liang, Z.H. Liu* and Y. Wang*, Development of human lactate dehydrogenase A inhibitors: High-throughput screening, molecular dynamics simulation and enzyme activity assay. J. Comput.-Aided Mol. Des. 38, 28 (2024) 171) J.D. Yue, Y. Yin, X. Feng, J. Xu, Y. Li, T. Li, S. Liang, X. He, Z.H. Liu* and Y. Wang*, Discovery of the Inhibitor Targeting the SLC7A11/xCT Axis through In Silico and In Vitro Experiments. International Journal of Molecular Sciences 25, 8284 (2024) 170) J.B. Zhu, P. Guo*, J.H. Zhang, Y.Z. Jiang, S.W. Chen, J.F. Liu, J. Jiang, J.G. Lan*, X.C. Zeng*, X. He* and J.R. Yang*, Superdiffusive rotation of interfacial water on noble metal surface. J. Am. Chem. Soc. 146, 16281 (2024) 169)Y. Zhang, H. Hao, X. He, S.H. Gao and A.M. Zhou*, Evolutionary Retrosynthetic Route Planning. IEEE Computational Intelligence Magazine 19, 58 (2024) 168) X. Liu#, J.C. Gong#, J.Z. Jiang, X. He* and J.R. Yang*, Optimizing Cell Voltage Dependence on Size of Carbon Nanotube-based Electrodes in Na-ion and K-ion batteries. Phys. Chem. Chem. Phys. 26, 12027 (2024) 167) H.T. Li#, M. Sun#, F.Z. Lei#, J.F. Liu, X.X. Chen, Y.Q. Li, Y. Wang, J.N. Lu, D.M. Yu, Y.Q. Gao, J.R. Xu, H.Z. Chen, M. Li*, Z.G. Yi*, X He* and L.L. Chen*, Methyl rosmarinate is an allosteric inhibitor of SARS-CoV-2 3CL protease as a potential candidate against SARS-CoV-2 infection, Antiviral Research 224, 105841 (2024) 166) J.D. Yue, J. Xu, Y. Yin, Y. Shu, Y. Li, T. Li, Z. Zou, Z. Wang, F. Li, M. Zhang, S. Liang, X. He, Z.H. Liu* and Y. Wang*, Targeting the PDK/PDH axis to reverse metabolic abnormalities by structure-based virtual screening with in vitro and in vivo experiments, International Journal of Biological Macromolecules 262, 129970 (2024) 165)W. Zhang#, C. Zhao#, W.J. Zhu, X. He* and Y.C. Zhao*, Conformational Locking as a Strategy to Reverse Ion Recognition Selectivity. J. Org. Chem. 89, 4037 (2024) 164) Y.N. Yang*#, S.W. Chen#, M. Zhang*, Y.R. Shi, J.Q. Luo, Y.M. Huang, Z.Y. Gu, W.L. Hu, Y. Zhang, X. He* and C.Z. Yu*, Mesoporous Nanoperforators as Membranolytic Agents via Nano- and Molecular- scale Multi-patterning. Nat. Commun. 15, 1891 (2024) 163) X.J. Zhou*, Z.R. Huang and X. He*, Diffusion Monte Carlo Method for Barrier Heights of Multiple Proton Exchanges and Complexation Energies in Small Water, Ammonia, and Hydrogen Fluoride Clusters. J. Chem. Phys. 160, 054103 (2024) 162) Z. Wan, M. Shi, Y.Q. Gong, M. Lucci, J.J. Li, J.H. Zhou, X.L. Yang*, M. Lelli*, X. He* and J.F. Mao*, Multitasking Pharmacophores Support Cabotegravir-Based 2 Long-Acting HIV Pre-Exposure Prophylaxis (PrEP). Molecules 29, 376 (2024) 161) G.X. Gu, C. Zhao, W. Zhang, J.J. Weng, Z.C. Xu, J. Wu, Y.B. Xie, X. He* and Y.C. Zhao*, Chiral Discrimination of Acyclic Secondary Amines By 19F NMR. Analytical Chemistry 96, 730 (2024) 160) J.F. Mao*, X.S. Jin, M. Shi, D. Heidenreich, L.J. Brown, R.C.D. Brown, M. Lelli, X. He* and C. Glaubitz*, Molecular Mechanisms and Evolutionary Robustness of a Color Switch in Proteorhodopsins. Sci. Adv. 10, eadj0384 (2024) 159) J.C. Gong, J.B. Zhu, X. He* and J.R. Yang*, Using Cyclocarbon Additive as Cyclone Separator to Achieve Fast Lithiation and Delithiation without Dendrite Growth in Lithium-ion Batteries. Nanoscale 16, 427 (2024) 158) Y. Lan, X. He*, X.M. Fang, L.H. Liu* and J.F. Liu*, Deep learning with geometry-enhanced molecularrepresentation for augmentation of ultralarge-scaledocking-based virtual screening. J. Chem. Inf. Model. 63, 6501 (2023) 157) C. Zhao, G.P. Yang, S. Zhang, X. He*, Y.S. Zhong and X.L. Gao*, Enhanced Breathing Effect of Nanoporous UIO-66-DABAMetal-Organic Frameworks with Coordination Defects forHigh Selectivity and Rapid Adsorption of Hg(II). ACS Applied Nano Materials 6, 18372 (2023) 156) J.H. Zhang, C. Kriebel, Z. Wan, M. Shi, C. Glaubitz* and X. He*, Automated Fragmentation Quantum Mechanical Calculation of 15N and 13C Chemical Shifts in a Membrane Protein J. Chem. Theory Comput. 19, 7405 (2023) 155) Y.Z. Jiang, Z. Wan, X. He* and J.R. Yang*, Fine-Tuning Metal-Organic Framework Electrolyte Interface toward Actuating Fast Zn2+ Dehydration for Aqueous Zn-Ion Batteries. Angew. Chem. Int. Ed. 62, e202307274 (2023) (Very Important Paper, VIP) 154) Y.T. Li, J.R. Yang* and X. He*, Characterizing polyproline II conformational change of collagen superhelix unit adsorption on gold surface. Nanoscale Advances 5, 5322 (2023) 153) E.D. Feng, T.T. Zheng*, X.X. He, J.Q. Chen, Q.Y. Gu, X. He, F.H. Hu, J.H. Li* and Y. Tian*, Plasmon-Induced Charge Transfer-Enhanced Raman Scattering on a Semiconductor: Toward Amplification-Free Quantification of SARS-CoV-2. Angew. Chem. Int. Ed. 62, e202309249 (2023) 152) S.Q. Li#, R.S. Peng#, Z. Wan, Y.D. Gong, X.M. Si, J. Tuo, H. Xu*, J.G. Jiang*, Y.J. Guan, Y.H. Ma, X. He* and P. Wu*, A Nanostrips-Assemble Morphology of ZSM-5 Zeolite for Efficient Propylene Production from Methanol Conversion. ACS Sustainable Chem. Eng. 11, 10274 (2023) 151) R.S. Peng#, S.Q. Li#, Z. Wan#, Z.Q. Wang, X.M. Si, J. Tuo, H. Xu, Y.J. Guan, J.G. Jiang*, Y.H. Ma, X. He*, X.Q. Gong* and P. Wu*, Directing Highly a-Axis Oriented ZSM-5 Nanosheets with Pre-Estimated Bifunctional Imidazole Cations. ACS Appl. Mater. Interfaces 15, 28116 (2023) 150) Y. Li, Y. Wang, R.M. Zhang, X. He* and X.F. Xu*, A Comprehensive Theoretical Study on Four Typical Intramolecular Hydrogen Shift Reactions of Peroxy Radicals: Multi-Reference Character, Recommended Model Chemistry, and Kinetics. J. Chem. Theory Comput. 19, 3284 (2023) 149) J.L. Wang#, Y.Q. Fan#, J.G. Jiang#, Z. Wan, S.Y. Pang, Y.J. Guan, H. Xu*, X. He*, Y.H. Ma*, A.S. Huang* and P. Wu*, Layered Zeolite for Assembly of Two-Dimensional Separation Membranes for Hydrogen Purification. Angew. Chem. Int. Ed. 62, e202304734 (2023) 148) L.L. Song#, L.L. Xiong#, D. Ni, W.X. Chen, J. Ji, J. Xue, X.W. Chen, X. Wu, X. He* and S.Y. Liu*, One-pot Construction of β-Selective Quinolines with γ- Quaternary Carbon from Vinylquinolines with Active Ylides via Pd/Sc/Brønsted Acid Co-Catalysis. ACS Catalysis 13, 6509 (2023) 147) Y. Wang#, Y.J. Qian#, L.M. Zhang*, Z.H. Zhang, S.W. Chen, J.F. Liu, X. He* and Y. Tian*, Conductive Metal−Organic Framework Microelectrodes Regulated by Conjugated Molecular Wires for Monitoring of Dopamine in the Mouse Brain. J. Am. Chem. Soc. 145, 2118 (2023) ESI 1% highly cited paper 146) Y.W. Liu, J.F. Liu* and X. He*, Different pKa Shifts of Internal GLU8 in Human β-endorphin Amyloid Reveal a Coupling of Internal Ionization and Stepwise Fibril Disassembly. J. Phys. Chem. B 127, 1089 (2023) (“Early-Career & Emerging Researchers in Physical Chemistry Volume 2”) 145)J.F. Liu and X. He*, Recent Advances in Quantum Fragmentation Approaches to Complex Molecular and Condensed-phase Systems. WIRES Comput. Mol. Sci. 13, e1650 (2023) 144)Y.W. Liu, C. Zhang, Z.H. Liu, D.G. Truhlar*, Y. Wang* and X. He*, Supervised learning of a chemistry functional with damped dispersion. Nat. Comput. Sci. 3, 48 (2023) 143) C. Zhang, P. Verma, J.X. Wang, Y.W. Liu, X. He, Y. Wang*, D.G. Truhlar* and Z.H. Liu*, Performance of screened-exchange functionals for band gaps and lattice constants of crystals. J. Chem. Theory Comput. 19, 311 (2023) 142) J.D. Yue, Y. Li, F. Li, P. Zhang, Y. Li, J. Xu, Q. Zhang, C. Zhang, X. He, Y. Wang* and Z.H. Liu*, Discovery of Mcl-1 inhibitors through virtual screening, molecular dynamics simulations and in vitro experiments, Computers in Biology and Medicine 152, 106350 (2023) 141) F. Wang, J.H. Zhang, M.D. Zhang, C.Y. Xu, S.Q. Cheng, Q.J. Wang, F. Zhang*, X. He* and P.G. He, A multi-calibration potentiometric sensing array based on diboronic acid- 2 PtAu/CNTs nanozyme for home monitoring of urine glucose. Analytica Chimica Acta 1237, 340598 (2023) 140) L.L. Xiong, X. He* and J.R. Yang*, Origin of Humidity Influencing the Excited State Electronic Properties of Silicon Quantum Dots based Light-emitting Diodes. Phys. Chem. Chem. Phys. 24, 28222 (2022) 139) Y. Wang, J.C. Gong, X.W. Wang, W.J. Li, X.Q. Wang, X. He*, W. Wang* and H.B. Yang*, Multistate Circularly Polarized Luminescence Switching through Stimuli-induced Co-conformation Regulations of Pyrene-functionalized Topologically Chiral [2]Catenane. Angew. Chem. Int. Ed. 61, e202210542 (2022) (Inside back cover)
138) W.N. Fang#, J.M. Wang#, S. Lu#, Q.Y. Gu, X. He, F. Wang, L.H. Wang, Y. Tian, H.J. Liu* and C.H. Fan, Encoding Morphogenesis of Quasi-Triangular Gold Nanoprisms with DNA. Angew. Chem. Int. Ed. 61, e202208688 (2022) 137) J. L. Wang#, Y.Q. Fan#, X.W. Guo#, Q.Y. Gu, J.G. Jiang, Y.J. Guan, X. He, Y.H. Ma, H. Xu* and P.Wu, Direct Synthesis and Delamination of Swollen Layered Ferrierite for the Reductive Etherification of Furfural. ChemCatChem 14, e202200535 (2022) 136) L.X. Liu, C.H. Luo, J.H. Zhang, X. He, Y. Shen, B. Yan, Y. Huang*, F. Xia and L. Jiang, Synergistic Effect of Bio-Inspired Nanochannels: Hydrophilic DNA Probes at Inner Wall and Hydrophobic Coating at Outer Surface for Highly Sensitive Detection. Small 18, 2201925 (2022) 135) W.Z. Yue#, Z. Wan#, Y.H. Li*, X. He*, J. Caro and A.S. Huang*, Synthesis of Cu-ZnO-Pt@HZSM-5 catalytic membrane reactor for CO2 hydrogenation to dimethyl ether. J. Membr. Sci. 660, 120845 (2022) 134) S.J. Chen#, X.Y. Ding#, C. Sun, F. Wang, X. He*, A. Watts* and X. Zhao*, Archaeal Lipids Regulating the Trimeric Structure Dynamics of Bacteriorhodopsin for Efficient Proton Release and Uptake. International Journal of Molecular Sciences 23, 6913 (2022) 133) T. Li, L. Yu, J.F. Sun*, J.F. Liu* and X. He*, The ionization of D571 is coupled with SARS-CoV-2 spike up/down equilibrium revealing the pH-dependent allosteric mechanism of receptor-binding domains. J. Phys. Chem. B 126, 4828 (2022) 132) C.H. Ma, Y. Quan, J.H. Zhang, R.Y. Sun, Q.H. Zhao, X. He, X.J. Liao* and M.R. Xie, Efficient Synthesis and Cyclic Molecular Topology of Ultra-Large Sized Bicyclic and Tetracyclic Polymers. Macromolecules 55, 4341 (2022) 131) J.F. Liu, J.G. Lan* and X. He*, Towards High-level Machine Learning Potential for Water based on Quantum Fragmentation and Neural Network. J. Phys. Chem. A 126, 3926 (2022) 130) Y.X. Mei, Z.C. Liu, M.J. Liu, J.C. Gong, X. He, Q.W. Zhang* and Y. Tian*, Two-photon fluorescence imaging and ratiometric quantification of mitochondrial monoamine oxidase-A in neurons. Chem. Comm. 58, 6657 (2022) 129) Y. Pan, J.X. Bao, X.Y. Zhang, H. Ni, Y. Zhao, F.D. Zhi, B.H. Fang, X. He*, J.Z.H. Zhang*, and L.J. Zhang*, Rational Design of P450 aMOx for improving anti-Markovnikov selectivity based on Butterfly Model. Frontiers in Molecular Biosciences 9, 888721 (2022) 128) Y.M. Li, Y.Q. Li, C. Ning, J.D. Yue, C. Zhang, X. He, Y. Wang* and Z.H. Liu, Discovering inhibitors of TEAD palmitate binding pocket through virtual screening and molecular dynamics simulation. Comput. Biol. Chem. 98, 107648 (2022) 127)A. Hu#, J.Z. Zhang#, J. Wang, C.C. Li, M. Yuan, G. Deng, Z.C. Lin, Z.P. Qiu, H.Y. Liu, X.W. Wang, P.C. Wei, X. He, X.L. Zhao*, W.W. Qiu* and B.L. Song*, Cholesterylation of Smoothened is a calcium-accelerated autoreaction involving an intramolecular ester intermediate. Cell Research 32, 288 (2022) 126) Y.X. Mei, Q.W. Zhang, Q.Y. Gu, Z.C. Liu, X. He* and Y. Tian*, Pillar[5]arene-Based Fluorescent Sensor Array for Biosensing of Intracellular Multi-Neurotransmitters through Host-Guest Recognitions. J. Am. Chem. Soc. 144, 2351 (2022) ESI 1% highly cited paper 125) H. Guo, Z. Wan, Y.H. Li, X. He and A.S. Huang*, Synthesis of graphene oxide membrane for separation of p-xylene and o-xylene by pervaporation. Chemie Ingenieur Technik, 94, 78 (2022) 124) N.F. Kleimeier#, Y.W. Liu#, A.M. Turner, L.A. Young, C.H. Chin, T. Yang*, X. He*, J.I. Lo, B.M. Cheng, R.I. Kaiser*, Excited State Photochemically Driven Surface Formation of Benzene from Acetylene Ices on Pluto and in the Outer Solar System. Phys. Chem. Chem. Phys. 24, 1424 (2022) 123) Y.L. Miao, H. Ni, X.Y. Zhang, F.D. Zhi, X. Long, X.P. Yang, X. He* and L.J. Zhang*, Investigating mechanism of sweetness-intensity differences through dynamic analysis of sweetener–T1R2–membrane systems. Food Chemistry 374, 131807 (2022) 122) S.J. Chen#, X.Y. Ding#, C. Sun, A. Watts, X. He* and X. Zhao*, Dynamic coupling of tyrosine 185 with the bacteriorhodopsin photocycle, as revealed by chemical shifts, assisted AF-QM/MM calculations and molecular dynamic simulations. International Journal of Molecular Sciences 22, 13587 (2021) 121) C.F. Shen, X.W. Wang* and X. He*, Fragment-based Quantum Mechanical Calculation of Excited-State Properties of Fluorescent RNAs. Front. Chem. 9, 801062 (2021) 120) J.F. Liu and X. He*, Ab Initio Molecular Dynamics Simulation of Liquid Water with Fragment-based Quantum Mechanical Approach under Periodic Boundary Conditions. Chinese Journal of Chemical Physics, 34, 761 (2021) (Part of Special Issue John Z.H. Zhang Festschrift for celebrating his 60th birthday) 119) X. He*, G.H. Li* and D.H. Zhang*, A Tribute to Prof. John Z.H. Zhang, Chinese Journal of Chemical Physics, 34 (2021) (Preface of Special Issue John Z.H. Zhang Festschrift for celebrating his 60th birthday) 118) A.S. Deng, X.T. Shen, Z. Wan, Y.H. Li*, S.Y. Pang, X. He*, J. Caro and A.S. Huang*, Elimination of Grain Boundary Defects in Zeolitic Imidazolate Framework ZIF-95 Membrane via Solvent-Free Secondary Growth. Angew. Chem. Int. Ed. 133, 25667 (2021) (Hot paper) 117) Y. Zhou, Q.Y. Gu, T.Z. Qiu, X. He, J.Q. Chen, R.J. Qi, R. Huang, T.T. Zheng* and Y. Tian*, Ultrasensitive Sensing of Volatile Organic Compounds Using a Cu-Doped SnO2-NiO p-n Heterostructure That Shows Significant Raman Enhancement. Angew. Chem. Int. Ed. 60, 26260 (2021) (Very Important Paper, VIP) 116) X.W. Wang*, X.L. Li, X. He* and J.Z.H. Zhang*, A Fixed Multi-Site Interaction Charge Model for an Accurate Prediction of the QM/MM Interactions. Phys. Chem. Chem. Phys. 23, 21001 (2021) 115) D.H. Dai#, X.W. Wang#, Y.W. Liu, X.L. Yang, C. Glaubitz, V. Denysenkov, X. He*, T. Prisner and J.F. Mao*, Room-Temperature dynamic nuclear polarization enhanced NMR Spectroscopy of Small Biological Molecules in Water. Nat. Commun. 12, 6880 (2021) 114) J.F. Liu#, J.R. Yang#, X.C. Zeng, S.S. Xantheas*, K. Yagi* and X. He*, Towards Complete Assignment of the Infrared Spectrum of the Protonated Water Cluster H+(H2O)21. Nat. Commun. 12, 6141 (2021) 113) C.F. Shen, X.S. Jin, W.J. Glover and X. He*, Accurate Prediction of Absorption Spectral Shifts of Proteorhodopsin Using a Fragment-Based Quantum Mechanical Method. Molecules 26, 4486 (2021) 112) X.X. Zhao#, P. Zhang#*, Y.Q. Li#, S.Z. Wu, F.J. Li, Y. Wang, S.P. Liang, X. He, Y.L. Zeng* and Z.H. Liu*, Glucose−Lipopeptide Conjugates Reveal the Role of Glucose Modification Position in Complexation and the Potential of Malignant Melanoma Therapy. J. Med. Chem. 64, 11483 (2021)
111) B. Long*, Y. Wang, Y. Xia, X. He*, J.L. Bao* and D.G. Truhlar*, Atmospheric Kinetics, Bimolecular Reactions of Carbonyl Oxide by a Triple-Level Strategy. J. Am. Chem. Soc. 143, 8402 (2021)
110) Z.S. Sun, Z.H. Gong, F. Xia and X. He*, Ion Dynamics and Selectivity of Nav channels from Molecular Dynamics Simulation. Chem. Phys. 548, 111245 (2021)
109) J.X. Bao, X. He* and J.Z.H. Zhang*, DeepBSP-A Machine Learning Method for Accurate Prediction of Protein-Ligand Docking Structure. J. Chem. Inf. Model. 61, 2231 (2021)
108) L.J. Zhang, J.K. Zheng, M.Z. Ma, Y. Zhao, J. Song, X. Chen, W.X. Cao, X. He, C.H. Xue and Q.J. Tang*, Drug-Guided Screening for Pancreatic Lipase Inhibitors in Functional Foods. Food & Function 12, 4644 (2021) 107) R.Y. Chen, Y.L. Miao, X. Hao, B. Gao, M.Z. Ma, J.Z.H. Zhang, R. Wang, S. Li, X. He* and L.J. Zhang*, Investigation on the characteristics and mechanisms of ACE inhibitory peptides by a thorough analysis of all 8000 tripeptides via binding free energy calculation. Food Science & Nutrition 9, 2943 (2021)
106) G.F. Shao, J.X. Bao, X.L. Pan, X. He*, Y.F. Qi* and J.Z.H. Zhang*, Analysis of the Binding Modes of the First- and Second-Generation Antiandrogens with respect to F876L Mutation. Chemical Biology & Drug Design 98, 60 (2021)
105) W.J. Li, Q.Y. Gu, X.Q. Wang, D.Y. Zhang, Y.T. Wang, X. He*, W. Wang,* and H.B. Yang, AIE-active Chiral [3]Rotaxanes with Switchable Circularly Polarized Luminescence. Angew. Chem. Int. Ed. 60, 9507 (2021) 104) X. Hao#, J.F. Liu#, I. Ali#, H.Y. Luo, Y.Q. Han, W.X. Hu, J.Y. Liu*, X. He* and J.J. Li*, Ab initio Determination of Crystal Stability of Di-p-tolyl Disulfide. Sci. Rep. 11, 4076 (2021)
103) J.F. Liu, Y.Q. Liu, J.R. Yang, X.C. Zeng and X. He*, Directional Proton Transfer in the Reaction of the Simplest Criegee Intermediate with Water Involving the Formation of Transient H3O+. J. Phys. Chem. Lett. 12, 3379 (2021)
102) M. Shi, J.S. Jin, Z. Wan and X. He*, Automated Fragmentation Quantum Mechanical Calculation of 13C and 1H Chemical Shifts in Molecular Crystals. J. Chem. Phys. 154, 064502 (2021) (2021 JCP Emerging Investigators Special Collection) 101) G.F. Shao, J.X. Bao, X.L. Pan, X. He*, Y.F. Qi* and J.Z.H. Zhang*, Computational Analysis of Residue-Specific Binding Free Energies of Androgen Receptor to Ligands. Frontiers in Molecular Biosciences, 8, 36 (2021) 100) J.X. Bao, X. He* and J.Z.H. Zhang*, Development of a New Scoring Function for Virtual Screen: APBScore. J. Chem. Inf. Model. 60, 6355 (2020) 99) X.X. Ma#, Z. Wan#, Y.H. Li, X. He*, J. Caro and A.S. Huang*, Anisotropic Gas Separation in Oriented ZIF-95 Membranes Prepared by Vapor-Assisted In-Plane Epitaxial Growth. Angew. Chem. Int. Ed. 59, 20858 (2020) (Very Important Paper, VIP) 98) B.H. Zhang, Y.J. Ma, X.S. Jin, Y. Wang, B.B. Suo*, X. He* and Z. Jin* GridMol2.0: Implementation and Application of Linear-scale Quantum Mechanics Methods and Molecular Visualization. Int. J. Quantum Chem. 120, e26402 (2020) 97) X.S. Jin, W.J. Glover* and X. He*, Fragment Quantum Mechanical Method for Excited States of Proteins: Development and Application to the Green Fluorescent Protein. J. Chem. Theory Comput., 16, 5174 (2020) 96) M. Kingsland, K.A. Lynch, S. Lisenkov*, X. He, and I. Ponomareva, Comparative Study of Minnesota Functionals Performance on Ferroelectric BaTiO3 and PbTiO3. Phys. Rev. Mater., 4, 073802 (2020) 95) J.F. Liu, and X. He*, Fragment-based Quantum Mechanical Approach to Biomolecules, Molecular 94) L. Huang, Y.Q. Han, J.Y. Liu*, X. He* and J.J. Li*, Ab Initio Prediction of the Phase Transition for Solid Ammonia at High Pressures. Sci. Rep., 10, 7546 (2020) 93) Z.L. Wang#, Y.Q. Han#, J.J. Li* and X. He*, Combining the Fragmentation Approach and Neural Network Potential Energy Surfaces of Fragments for Accurate Calculation of Protein Energy. J. Phys. Chem. B, 124, 3027 (2020) 92) W. Zhang, J.F. Liu, X.S. Jin, X.G. Gu*, X.C. Zeng, X. He* and H. Li*, Quantitative Prediction of Aggregation-Induced Emission: A Full Quantum Mechanical Approach to the Optical Spectra. Angew. Chem. Int. Ed. 59, 11550 (2020) 91) J.J. Xu#, J.F. Liu#, J.Y. Liu*, W.X. Hu, X. He* and J.J. Li*, Phase Transition of Ice at High Pressures and Low Temperatures. Molecules, 25, 486 (2020) 90) Y. Wang, P. Verma, L.J. Zhang, Y.Q. Li, Z.H. Liu, D.G. Truhlar* and X. He*, M06-SX Screened-Exchange Density Functional for Chemistry and Solid-State Physics. Proc. Natl. Acad. Sci. U.S.A., 117, 2294 (2020) 89) J.F. Liu and X. He*, QM Implementation in Drug Design: Does It Really Help? Book chapter, Quantum Mechanics in Drug Discovery. in Methods in Molecular Biology, 2114, 19-35 (2020) 88) L. Huang, Y.Q. Han, X. He* and J.J. Li*, Ab initio-enabled Phase Transition Prediction of Solid Carbon Dioxide at Ultra-high Temperatures. Rsc Adv., 10, 236 (2020) 87) Y.W. Wu, H.Y. Mu, X.J. Cao* and X. He*, Polymer Supported Graphene-TiO2 Doped with Non-metallic Elements with Enhanced Photocatalytic Reaction under Visible Light. Journal of Materials Science, 55, 1577 (2020) 86) J.F. Mao*, V. Aladin, X.S. Jin, A.J. Leeder, L.J. Brown, R.C.D. Brown, X. He, B. Corzilius and C. Glaubitz*, Exploring Protein Structures by DNP-Enhanced Methyl Solid-State NMR Spectroscopy. J. Am. Chem. Soc., 141, 19888 (2019)
85) W.Z. Cao, D.J. Mei*, Y. Yang, Y.W. Wu, L.Y. Zhang, Y.D. Wu, X. He, Z.S. Lin,* and F.Q. Huang*, From CuFeS2 to Ba6Cu2FeGe4S16: Rational Band Gap Engineering Achieves Large Second-Harmonic-Generation Together with High Laser Damage Threshold. Chem. Comm., 55, 14510 (2019) 84) X.S. Jin and X. He*, Calculation of Biomolecular Nuclear Magnetic Resonance Chemical Shift Based on the Fragmentation Quantum Chemical Method. Journal of Technology, 19, 2096 (2019) 83) H.Y. Luo, J.Y. Liu*, X. He* and J.J. Li*, Low-Temperature Polymorphic Transformation of β-Lactam Antibiotics. Crystals, 9, 460 (2019) 82) P. Verma, B.G. Janesko*, Y. Wang, X. He, G. Scalmani, M.J. Frisch and D.G. Truhlar*, M11plus: A Range-Separated Hybrid Meta Functional with Both Local and Rung-3.5 Correlation Terms and High Across-the-Board Accuracy for Chemical Applications. J. Chem. Theory Comput., 15, 4804 (2019) (Cover article)
81) J.F. Liu, H.T. Sun, W.J. Glover and X. He*, Prediction of Excited-State Properties of Oligoacene Crystals Using Fragment-Based Quantum Mechanical Method. J. Phys. Chem. A, 123, 5407 (2019) 80) X. Hao#, J.F. Liu#, H.Y. Luo, Y.Q. Han, W.X. Hu, J.Y. Liu*, J.J. Li* and X. He*, Crystal Structure Optimization and Gibbs Free Energy Comparison of Five Sulfathiazole Polymorphs by the Embedded Fragment QM Method at the DFT Level. Crystals, 9, 256 (2019) 79) Q.N. Lu#, X. He#,W.X. Hu, X.J. Chen* and J.F. Liu*, Stability, Vibrations, and Diffusion of Hydrogen Gas in Clathrate Hydrates: Insights from Ab Initio Calculations on Condensed-Phase Crystalline Structures. J. Phys. Chem. C, 123, 12052 (2019) 78) Y.Q. Han#, J.F. Liu#, L. Huang, X. He* and J.J. Li*, Predicting the phase diagram of solid carbon dioxide at high pressure from first principles, NPJ Quantum Materials, 4, 10 (2019) 77) H.Y. Luo, X. Hao, Y.Q. Gong, J.H. Zhou, X. He* and J.J. Li*, Rational Crystal Polymorph Design of Olanzapine, Cryst. Growth Des., 19, 2388 (2019) 76) P. Verma*, Y. Wang, S. Ghosh, X. He* and D.G. Truhlar*, Revised M11 Exchange–Correlation Functional for Electronic Excitation Energies and Ground–State Properties. J. Phys. Chem. A, 123, 2966 (2019) 75) L.Y. Zhang, D.J. Mei*, Y.W. Wu, C.F. Shen W.X. Hu, L.J. Zhang, J.J. Li, Y.D. Wu and X. He, Syntheses, structures, optical properties, and electronic structures of Ba6Cu2GSn4S16 (G = Fe, Ni) and Sr6D2FeSn4S16 (D = Cu, Ag). J. Solid State Chem., 272, 69 (2019) 74) M.Y. Xu, X. He, T. Zhu* and J.Z.H. Zhang*, A Fragment Quantum Mechanical Method for Metalloproteins. J. Chem. Theory Comput., 15, 1430 (2019) 73) J.F. Liu, J.Z.H. Zhang* and X. He*, Probing the Ion-Specific Effects at the Water/Air Interface and Water-Mediated Ion Pairing in Sodium Halide Solution with Ab initio Molecular Dynamics. J. Phys. Chem. B 122, 10202 (2018) 72) X.W. Wang and X. He*, An Ab Initio QM/MM Study of the Electrostatic Contribution to Catalysis in the Active Site of Ketosteroid Isomerase. Molecules, 23, 2410 (2018) 71) Y. Wang#, P. Verma#, X.S. Jin, D.G. Truhlar* and X. He*, Revised M06 Density Functional for Main-Group and Transition- Metal Chemistry. Proc. Natl. Acad. Sci. U.S.A., 115, 10257 (2018) 70) X.Y. Ding, C. Sun, H.L. Cui, S.J. Chen, Y.J. Gao, Y.N. Yang, J. Wang, X. He, D. Luga, F. Tian*, A. Watts* and X. Zhao*, Functional Roles of Tyrosine 185 During the Bacteriorhodopsin Photocycle as Revealed by in situ Spectroscopic Studies. Biochimica et Biophysica Acta (BBA) - Bioenergetics, 1859, 1006 (2018) 69) Z.Q. Yao, L.J. Zhang*, B. Gao, D.B. Cui, F.Q. Wang, X. He, J.Z.H. Zhang and D.Z. Wei*, Crius: A Novel Fragment-based Algorithm of De Novo Substrate Prediction for Enzymes. Protein Science, 27, 1526 (2018) 68) X.S. Jin, T. Zhu, J.Z.H. Zhang and X. He*, Automated Fragmentation QM/MM Calculation of NMR Chemical Shifts for Protein-Ligand Complexes. Front. Chem., 6, 150 (2018) 67) Y.Q. Wang, J.F. Liu*, J.J. Li* and X. He*, Fragment-based Quantum Mechanical Calculation of Protein-protein Binding Affinities. J. Comput. Chem., 39, 1617 (2018) 66) B. Zhou, Z.B. Hu, X. He, Z.R. Sun and H.T. Sun*, Benchmark Study of Ionization Potentials and Electron Affinities of Single-Walled Carbon Nanotubes using Denisty Functional Theory. J. Phys. Condens. Matter., 30, 215501, (2018) 65) J.F. Liu, J. Swails, J.Z.H. Zhang, X. He* and A.E. Roitberg*, A Coupled Ionization-Conformational Equilibrium Is Required To Understand The Properties of Ionizable Residues in the Hydrophobic Interior of Staphylococcal Nuclease. J. Am. Chem. Soc., 140, 1639 (2018) 64) J.F. Liu#, X. He#, J.Z.H. Zhang and L.W. Qi*, Hydrogen-bond Structure Dynamics in Bulk Water: Insights from ab initio Simulations with Coupled Cluster Theory. Chem. Sci., 9, 2065 (2018) (Inside front cover)
63) Y. Wang, X.W. Wang, D.G. Truhlar* and X. He*, How Well Can the M06 Suite of Functionals Describe the Electron Densities of Ne, Ne6+, and Ne8+? J. Chem. Theory Comput., 13, 6068 (2017) 62) J.L. Bao, Y. Wang, X. He, L. Gagliardi* and D.G. Truhlar*, Multiconfiguration Pair-Density Functional Theory Is Free From Delocalization Error. J. Phys. Chem. Lett., 8, 5616 (2017)
60) Y. Zhang* and X. He, Reaction mechanisms of CO oxidation on cationic, neutral, and anionic X-O-Cu (X = Au, Ag) clusters. Chem. Phys. Lett., 686, 116 (2017) 59) Y. Wang, J.F. Liu, L.J. Zhang, X. He* and J.Z.H. Zhang*, Computational Search for Aflatoxin Binding Proteins. Chem. Phys. Lett., 685, 1 (2017) 58) J.F. Liu and X. He*, Accurate Prediction of Energetic Properties of Ionic Liquid Clusters Using Fragment-based Quantum Mechanical Method. Phys. Chem. Chem. Phys., 19, 20657 (2017) 57) Y. Wang, X.S. Jin, H.S. Yu, D.G. Truhlar* and X. He*, Revised M06-L Functional for Improved Accuracy on Chemical Reaction Barrier Heights, Noncovalent Interactions, and Solid-state Physics. Proc. Natl. Acad. Sci. U.S.A., 114, 8487 (2017) 56) S. Hirata, K. Gilliard, X. He, M. Keceli, J.J. Li, M.A. Salim, O. Sode and K. Yagi, Ab initio ice, dry ice and liquid water. (Chapter 9 of the book "Fragmentation: Toward Accurate Calculations on Complex Molecular Systems" edited by Prof. Mark Gordon, 2017) 55) J.F. Liu, T. Zhu, X. He and J.Z.H. Zhang, MFCC Based Fragmentation Methods for Biomolecules. (Chapter 11 of the book "Fragmentation: Toward Accurate Calculations on Complex Molecular Systems" edited by Prof. Mark Gordon, 2017) 54) J.F. Liu, X. He* and J.Z.H. Zhang*, Structure of Liquid Water - A Dynamical Mixture of Tetrahedral and ‘Ring-and-Chain’ like Structures. Phys. Chem. Chem. Phys., 19, 11931 (2017) 53) J.F. Liu, L.W. Qi*, J.Z.H. Zhang and X. He*, Fragment Quantum Mechanical Method for Large-sized Ion-water Clusters. J. Chem. Theory Comput., 13, 2021 (2017) 52) X.S. Jin, J.Z.H. Zhang and X. He*, Full QM Calculation of RNA Energy Using Electrostatically Embedded Generalized Molecular Fractionation with Conjugate Caps Method. J. Phys. Chem. A, 121, 2503 (2017) 51) X.S. Jin, T. Zhu, J.Z.H. Zhang and X. He*, A Systematic Study on RNA NMR Chemical Shift Calculation Based on the Automated Fragmentation QM/MM Approach. RSC Advances, 6, 108590 (2016) 50) J.F. Liu, Y.Q. Wang, J.Z.H. Zhang and X. He*, Quantum mechanical mechanism of binding of 4-Anilinoquinazoline inhibitors to the Epidermal Growth Factor Receptor based on MFCC computation. China Science Paper, 11, 2050 (2016) 49) J.W. Yuan, J.P. Ren, Y. Wang, X. He and Y.W. Zhao*, Acteoside Binds to Caspase-3 and Exerts Neuroprotection in the Rotenone Rat Model of Parkinson's Disease. PLoS ONE, 11, e0162696 (2016) 48) Z.Q. Yao, L.J. Zhang*, B. Gao, D.B. Cui, F.Q. Wang, X. He, J.Z.H. Zhang and D.Z. Wei*, A Semiautomated Structure-Based Method to Predict Substrates of Enzymes via Molecular Docking: A Case Study with Candida Antarctica Lipase B. J. Chem. Inf. Model., 56, 1979 (2016) 47) X.W. Wang, X. He* and J.Z.H. Zhang*, Accurate calculation of electric fields inside enzymes. Methods in Enzymology, 578, 45 (2016) 46) Y. Wang, J.F. Liu, T. Zhu, L.J. Zhang, X. He* and J.Z.H. Zhang*, Predicted PAR1 inhibitors from multiple computational methods. Chem. Phys. Lett., 659, 295 (2016) 45) X. Liu, J.F. Liu, T. Zhu, L.J. Zhang, X. He* and J.Z.H. Zhang*, PBSA_E: A PBSA-Based Free Energy Estimator for Protein-Ligand Binding Affinity. J. Chem. Inf. Model., 56, 854 (2016) 44) H.S. Yu, X. He, S.L. Li and D.G. Truhlar*, MN15: A Kohn–Sham Global-Hybrid Exchange-Correlation Density Functional with Broad Accuracy for Multi-Reference and Single-Reference Systems and Noncovalent Interactions. Chem. Sci., 7, 5032 (2016) ESI 1% highly cited paper 43) Y.X. Li, S.Q. Zhang, J.Z.H. Zhang and X. He*, Assessing the Performance of Popular QM Methods for Calculation of Conformational Energies of Trialanine. Chem. Phys. Lett. 652, 136 (2016) 42) H.S. Yu, X. He and D.G. Truhlar*, MN15-L: A New Local Exchange-Correlation Functional for Kohn–Sham Density Functional Theory with Broad Accuracy for Atoms, Molecules and Solids. J. Chem. Theory Comput. 12, 1280 (2016) ESI 1% highly cited paper 41) J.F. Liu, J.Z.H. Zhang and X. He*, Fragment Quantum Chemical Approach to Geometry Optimization and Vibrational Spectrum Calculation of Proteins. Phys. Chem. Chem. Phys. 18, 1864 (2016) 40) P.C. Shen, W.W. Li, Y. Wang, X. He and L.Q. He*, Binding Mode of Chitin and TLR2 via Molecular Docking and Dynamics Simulation. Molecular Simulation, 42, 936 (2016) 39) J.F. Liu, X.W. Wang, J.Z.H. Zhang and X. He*, Calculation of Protein-Ligand Binding Affinities Based on Fragment Quantum Mechanical Method. RSC Advances, 5, 107020 (2015) 38) J.F. Liu, T. Zhu*, X.W. Wang, X. He* and J.Z.H. Zhang*, Quantum Fragment Based ab Initio Molecular Dynamics for Proteins. J. Chem. Theory Comput. 11, 5897 (2015) 37) X.W. Wang, J.Z.H. Zhang and X. He*, Quantum Mechanical Calculation of Electric Fields and Vibrational Stark Shifts at the Active Site of Human Aldose Reductase. J. Chem. Phys. 143, 184111 (2015) 36)J. Swails, T. Zhu, X. He* and D.A. Case*, AFNMR: Automated Fragment Quantum mechanical Calculation of NMR Chemical shifts for Biomolecules. Journal of Biomolecular NMR, 63, 125 (2015) 35)H.S. Yu, W.J. Zhang, P. Verma, X. He and D.G. Truhlar*, Nonseparable Exchange-Correlation Functional for Molecules, Including Homogeneous Catalysis Involving Transition Metals. Phys. Chem. Chem. Phys. 17, 12146 (2015) 34)T. Zhu, J.Z.H. Zhang and X. He*, Quantum Calculation of Protein NMR Chemical Shifts Based on the Automated Fragmentation Method. (Advance in Structural Bioinformatics, Advances in Experimental Medicine and Biology, 827, 49, Springer, 2015, IF=2.012) 33) J.F. Liu, X. He* and J.Z.H. Zhang, Novel Theoretically Designed HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitors Derived from Nevirapine. J. Mol. Model., 20, 2451 (2014) 32) T. Zhu, J.Z.H. Zhang and X. He*, Correction of Erroneously Packed Protein's Side Chain in the NMR Structure based on ab initio chemical shift calculations. Phys. Chem. Chem. Phys., 16, 18163 (2014) 31)X. He*, T. Zhu, X.W. Wang, J.F. Liu and J.Z.H. Zhang*, Fragment Quantum Mechanical Calculation of Proteins and Its Applications. Acc. Chem. Res. 47, 2748 (2014) 30)S. Hirata*, K. Gilliard, X. He, J.J. Li and O. Sode, Ab Initio Molecular Crystal Structures, Spectra, and Phase Diagrams. Acc. Chem. Res. 47, 2721 (2014) 29) X.W. Wang, Y.X. Li, X. He*, S.D. Chen and J.Z.H. Zhang*, Effect of Strong Electric Field on Conformational Integrity of Insulin. J. Phys. Chem. A 118, 8942 (2014) 28)L.J. Zhang, B. Gao, Z.N. Yuan, X. He, Y.A. Yuan, J.Z.H. Zhang* and D.Z. Wei*, Structure, mechanism, and enantioselectivity shifting of lipase LipK107 with a simple way. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, 1844, 1183, (2014) (Impactor factor=3.733) 27)S. Hirata*, X. He, M.R. Hermes and S.Y. Willow, Second-order Many-body Perturbation Theory: An Eternal Frontier. J. Phys. Chem. A 118, 655 (2014) [Invited Feature Article] 26)X. He, S. Ryu and S. Hirata*, Finite-temperature second-order many-body perturbation and Hartree–Fock theories for one-dimensional solids: An application to Peierls and charge-density-wave transitions in conjugated polymers. J. Chem. Phys. 140, 024702 (2014) 25)X.Y. Jia, X.W. Wang, J.F. Liu, J.Z.H. Zhang, Y. Mei* and X. He*, An Improved Fragment-based Quantum Mechanical Method for Calculation of Electrostatic Solvation Energy of Proteins. J. Chem. Phys. 139, 214104 (2013) 24)B. Wang, X. He* and K.M. Merz*, Quantum Mechanical Study of Vicinal J Spin-Spin Coupling Constants for the Protein Backbone. J. Chem. Theory Comput. 9, 4653 (2013) 23)S. Hirata* and X. He, On the Kohn-Luttinger Conundrum.J. Chem. Phys. 138, 204112 (2013) 22)J.F. Liu, X. He* and J.Z.H. Zhang*, Improving the Scoring of Protein-ligand Binding Affinity by Including the Effects of Structural Water and Electronic Polarization. J. Chem. Inf. Model. 53, 1306 (2013) 21)J. Bao, J.F. Liu, X. He* and J.Z.H. Zhang, Computational Study of HIV-1 gp41 NHR trimer: Inhibition Mechanisms of N-Substituted Pyrrole Derivatives and Fragment-Based Virtual Screening. J. Theor. Comput. Chem. 12, 1341001 (2013) 20)Y.X. Li, Y. Gao, X.Q. Zhang, X.Y. Wang, L.R. Mou, L.L. Duan, X. He*, Y. Mei* and J.Z.H. Zhang*, A Coupled Two-dimensional Main Chain Torsional Potential for Protein Dynamics: Generation and Implementation. J. Mol. Model. 19, 3647 (2013) 19)X.W. Wang, X. He* and J.Z.H. Zhang*, Predicting Mutation-induced Stark Shifts in the Active Site of a Protein with a Polarized Force Field. J. Phys. Chem. A 117, 6015 (2013) 18)X.W. Wang, J.F. Liu, J.Z.H. Zhang and X. He*, Electrostatically Embedded Generalized Molecular Fractionation with Conjugate Caps Method for Full Quantum Mechanical Calculation of Protein Energy. J. Phys. Chem. A 117, 7149 (2013) 17) T. Zhu, J.Z.H. Zhang and X. He*, Automated Fragmentation QM/MM Calculation of Amide Proton Chemical Shifts in Proteins with Explicit Solvent Model. J. Chem. Theory Comput. 9, 2104 (2013) 16) X. He, O. Sode, S. S. Xantheas and S. Hirata*, Second-order Many-body Perturbation Study of Ice Ih. J. Chem. Phys. 137, 204505 (2012) 15) Y. Mei, X. He, C.G. Ji, D.W. Zhang and J.Z.H. Zhang*, A Fragmentation Approach to Quantum Calculation of Large Molecular Systems. Progress in Chemistry 24, 1058 (2012) 14) T. Zhu, X. He* and J.Z.H. Zhang, Fragment Density Functional Theory Calculation of NMR Chemical Shift for Proteins with Implicit Solvation. Phys. Chem. Chem. Phys. 14, 7837 (2012) 13) J. Faver, M. Benson, X. He, B. Roberts, B. Wang, M.S. Marshell, C.D. Sherrill and K.M. Merz*, The Energy Computation Paradox and ab initio Protein Folding. PLoS ONE, 6(4), e18868 (2011) 12) J. Faver, M. Benson, X. He, B. Roberts, B. Wang, M.S. Marshell, M.R. Kennedy, C.D. Sherrill and K.M. Merz*, Formal Estimation of Errors in Computed Absolute Interaction Energies of Protein-Ligand Complexes. J. Chem. Theory Comput., 7, 790 (2011) 11) X. He and K.M. Merz*, Divide and Conquer Hartree-Fock Calculations on Proteins. J. Chem. Theory Comput. 6, 405 (2010) 10) X. He, L. Fusti-Molnar and K.M. Merz*, Accurate Benchmark Calculations on the Gas-Phase Basicities of Small Molecules. J. Phys. Chem. A 113, 10096 (2009) 9) L. Fusti-Molnar, X. He, B. Wang and K.M. Merz*, Further Analysis and Comparative Study of the Intermolecular Interactions Using Dimers from the S22 Database. J. Chem. Phys. 131, 065102 (2009) 8) X. He, B. Wang and K.M. Merz*, Protein NMR Chemical Shift Calculations Based on the Automated Fragmentation QM/MM Approach. J. Phys. Chem. B 113, 10380 (2009) 7) X. Li, X. He, B. Wang and K.M. Merz*, Conformational Variability of Benzamidinium Based Inhibitoirs. J. Am. Chem. Soc. 131, 7742 (2009) 6) X. He, L. Fusti-Molnar, G. Cui and K.M. Merz*, The Importance of Dispersion and Electron Correlation in ab initio Protein Folding. J. Phys. Chem. B 113, 5290 (2009) 5) X. He and J.Z.H. Zhang*, The generalized molecular fractionation with conjugate caps/ molecular mechanics method for direct calculation of protein energy. J. Chem. Phys. 124, 184703 (2006) 4) X. He, Y. Mei, Y. Xiang, D.W. Zhang and J.Z.H. Zhang*, Quantum Computational Analysis for Drug Resistance of HIV-1 Reverse Transcriptase to Nevirapine through Point Mutations, PROTEINS 61, 423-432 (2005) 3) X. He and J.Z.H. Zhang*, A New Method for Direct Calculation of Total Energy of Protein, J. Chem. Phys. 122, 031103 (2005). 2) Y. Mei, X. He, Y. Xiang, D.W. Zhang and J.Z.H. Zhang*, Quantum Study of Mutational Effect in Binding of Efavirenz to HIV-1 RT, PROTEINS 59, 489-495 (2005) 1) Q. Cui,X. He, M.L. Wang and J.Z.H. Zhang*, Comparison of Quantum and Mixed Quantum-classical Semirigid Vibrating Rotor Target Studies for Isotopic Reactions H(D,T) + CH4 ---> HH(D,T) + CH3, J. Chem. Phys. 119, 9455-9460 (2003) 专利 2015,何晓*,刘金峰,张增辉,一种非核苷类HIV-1反转录酶抑制剂。(专利号:201310242563.7) 2020, 潘月,张鲁嘉,张增辉,何晓,张传玺,方波欢,一种基于酵母二肽基肽酶Ⅲ的抗体模拟物及其应用。(专利号:ZL202010192357.X) 2020,赵玥,张鲁嘉,张增辉,何晓,方波欢,一种基于α螺旋融合两种蛋白质且保持各自亚基活性的蛋白质融合设计方法。(专利号:ZL202010192367.3) 2020,赵玥,张鲁嘉,张增辉,韩艳芳,何晓,方波欢,一种表达微管β亚基与蛋白A的D结构域融合蛋白的基因工程菌株及其构建方法。(专利号:CN202010207523.9) 2022,刘顺英,宋龙龙,倪丹,何晓,鲍劲霄,熊露露,薛建,纪健,陈绪文,一种多取代喹啉衍生物及其合成方法和应用 (专利号: CN202210875559.3) 2023,高蓓,张鲁嘉,朱屹婷,冯英慧,王嘉伟,何晓,一种高酯化能力的脂肪酶突变体及其表达应用(专利号:CN202310560479.3) 2024, 张焱,郝昊,何晓,周爱民, 一种分子逆合成路线规划方法及规划系统。(专利号: 202411253711.X) 2025,何晓,胡连瑞,杨学明,陈欣梦,张跃庆,王飞,自动化反应系统。(专利号: 2024232898737) 软件著作权 2024, 分子逆合成系统软件,2024SR1583841 2024, 材料计算机器人软件,2024SR1072893 2024, 全自动分子对接软件,2024SR0418692 2017, 蛋白-蛋白结合自由能的精确量子计算软件平台,2019SR0035529 SOFTWARES 6) AIMD interfaced with AMBER, Jinfeng Liu, Tong Zhu, John Z.H. Zhang and Xiao He 5) SHIFTS5.0, Xiaoping Xu, Seongho Moon, Jason Swails, Tong Zhu, Xiao He and David A. Case 4) QUICK Xiao He, Ken Ayers, Ed Brothers and Kenneth M. Merz, Jr. Ab initio Quantum Chemistry Package 3) AF-NMR Xiao He and Kenneth M. Merz, Jr. Protein NMR Chemical Shift Calculations Based on the Automated Fragmentation QM/MM Approach 2) EDISON2.0 Xiao He, Yun Xiang, John Z.H. Zhang Protein Optimization in Explicit Water Box by Combined Quantum Mechanics and Molecular Force Field (GMFCC/MM) in MPI Version C 1) MQC-TINKER Xiao He, Ye Mei, Ming L. Wang and John Z.H. Zhang Mixed Quantum-classical Study of Energy Transfer in Ion Collision with Proteins 荣誉及奖励2024 科技部科技评估中心重点领域咨询专家 2024 高管项目最受学员欢迎教师奖 2024 高性能计算应用“戈登贝尔奖”入围 2024 CCF HPC China2024超算年度最佳应用奖 2024 华东师范大学理工科“2023年度十大科技进展” 2023 教育部“长江学者”特聘教授 2023 美国化学会优秀导师奖 2023 华东师范大学研究生教育卓越育人奖(优秀导师奖) 2019 中国化学会“中国青年化学家元素周期表”氪元素代言人 2019 华东师范大学紫江优秀青年学者 2019 中国化学会唐敖庆理论化学青年奖 2019 国家自然科学基金委优秀青年基金 2016 上海市普陀区青年英才 2016 上海市青年拔尖人才 2009 美国佛罗里达大学Crow-Stasch优秀科学论文奖 2005 南京大学优秀研究生奖学金 2002 全国大学生数学建模比赛江苏省赛区一等奖 2001 全国大学生数学建模比赛全国二等奖 1999-2001 南京大学人民奖学金 |

|
何晓 |