50. Xiao M., Wang X., Yao Q., Li L., Fan C., Pei H*. Boosting selective fusion of protocells with DNA logic circuits for in situ detection of exosomal microRNA. Chem. 2024
49. Wei J.+, Sun Y.+, Wang H.+, Zhu T.+, Li L.+, Zhou Y., Liu Q., Dai Z., Li W., Yang T., Wang B., Zhu C., Shen X.*, Yao Q.*, Song G.*, Zhao Y.*, Pei H*. Designer cellular spheroids with DNA origami for drug screening. Science Advances. 2024, 10(29), eado9880.
48. Ji W.+, Xiong X.+, Cao M.+, Zhu Y.+, Li L., Wang F., Fan C.*, Pei H*. Encoding signal propagation on topology-programmed DNA origami. Nature Chemistry. 2024, 16(9), 1408-1417.
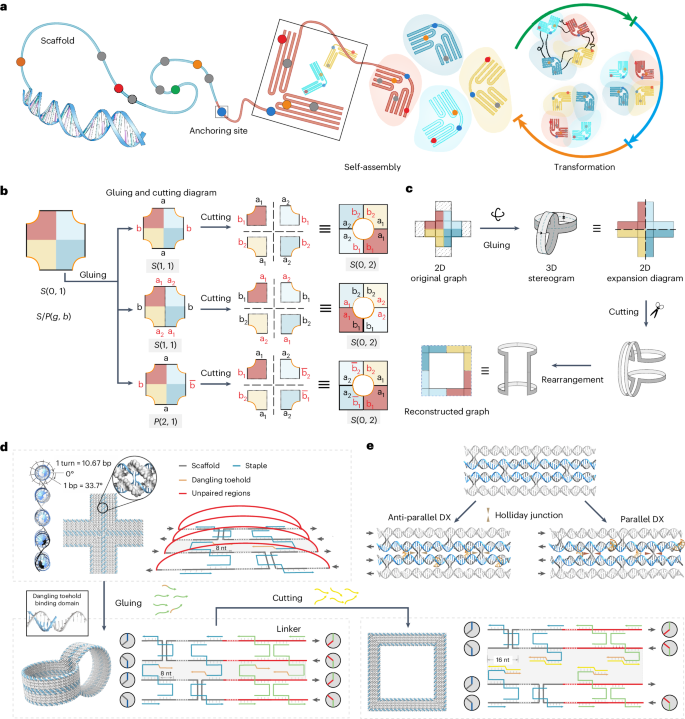
47. Wang L.+, Li N.+, Cao M., Zhu Y., Xiong X., Li L., Zhu T.*, Pei H*. Predicting DNA Reactions with a Quantum Chemistry-Based Deep Learning Model. Advanced Science. 2024, 2409880.
46. Wang L., Zhao J., Xiong X., Li L., Zhu T.*, Pei H*. Enzyme-Free Nucleic Acid Circuits for Fold-Change Detection. ChemPlusChem. 2023, 88(9), e202300083.
45. Xuan J., Wang Z., Xiao M.*, Pei H*. Engineering of Interfaces with Tetrahedra DNA Nanostructures for Biosensing Applications. Analysis & Sensing. 2023, 3(5), e202200100.
44. Zhu Y., Xiong X., Cao M., Li L., Fan C., Pei H*. Accelerating DNA computing via freeze-thaw cycling. Science Advances. 9(34), eaax7983.
43. Cao M., Xiong X., Zhu Y., Xiao M., Li L.*, Pei H*. DNA computational device-based smart biosensors. TrAC Trends in Analytical Chemistry. 2023, 159, 116911.
42. Sun Y., Sun J., Xiao M., Lai W., Li L.*, Fan C., Pei H.*, DNA origami-based artificial antigen-presenting cells for adoptive T cell therapy. Sci. Adv. 2022, 8(48), eadd1106.

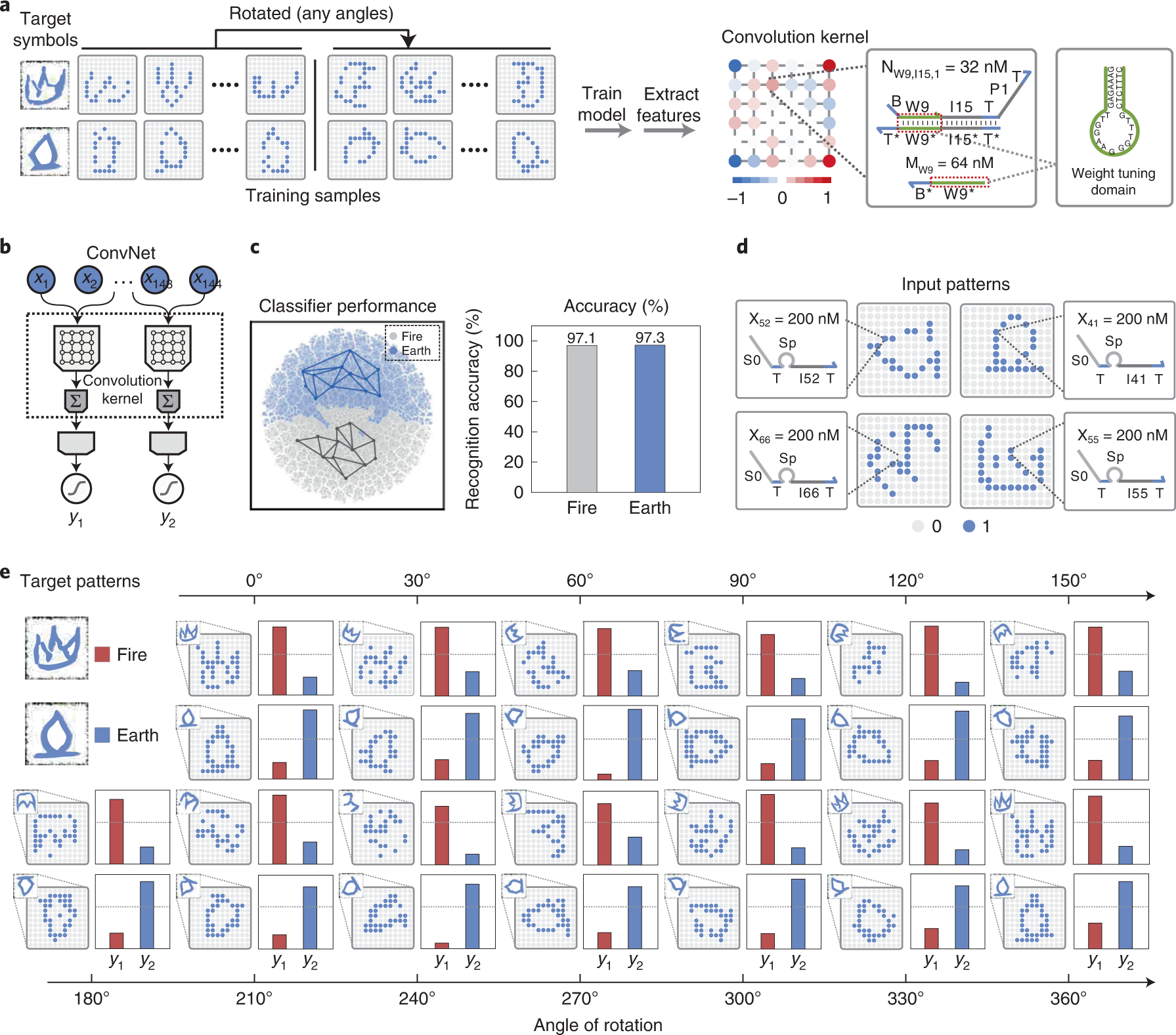
41. Xiong X.+, Zhu T.+, Zhu Y., Cao M., Xiao J., Li L., Wang F., Fan C., Pei H.* Molecular convolutional neural networks with DNA regulatory circuits. Nat. Mach. Intell. 2022, 4(7), 625-635.
40. Sun Y., Yan L., Sun J., Xiao M., Lai W., Song G., Li L., Fan C., Pei H.* Nanoscale organization of two-dimensional multimeric pMHC reagents with DNA origami for CD8(+) T cell detection. Nat. Commun. 2022, 13(1), 3916.
39. Shi S., Chen J., Wang X., Xiao M.*, Chandrasekaran A.R., Li L., Yi C.*, Pei H.* Biointerface Engineering with Nucleic Acid Materials for Biosensing Applications. Adv. Funct. Mater. 2022, 32(37).
38. Wang X., Yan L., Yu Z., Chen Q., Xiao M., Liu X., Li L.*, Pei H.* Aptamer-Functionalized Fractal Nanoplasmonics-Assisted Laser Desorption/Ionization Mass Spectrometry for Metabolite Detection. ChemPlusChem. 2022, 87(1), e202100479.
37. Wu Z., Xiao M., Lai W., Sun Y., Li L., Hu Z.*, Pei H.* Nucleic Acid-Based Cell Surface Engineering Strategies and Their Applications. ACS Appl. Bio. Mater. 2022, 5(5), 1901-1915.
36. Song L., Xiao M., Lai W., Li L., Wan Y., Pei H.* Intracellular Logic Computation with Framework Nucleic Acid‐Based Circuits for mRNA Imaging. Chin. J. Chem. 2021, 39(4), 947-953.
35. Tang Q.+, Lai W.+, Wang P., Xiong X., Xiao M., Li L., Fan C., Pei H.* Multi-Mode Reconfigurable DNA-Based Chemical Reaction Circuits for Soft Matter Computing and Control. Angew. Chem. Int. Ed. 2021, 60(27), 15013-15019.
34. Xiao M.+, Lai W.+, Yu H., Yu Z., Li L., Fan C., Pei H.* Assembly Pathway Selection with DNA Reaction Circuits for Programming Multiple Cell-Cell Interactions. J. Am. Chem. Soc. 2021, 143(9), 3448-3454.
33. Xiong X., Xiao M., Lai W., Li L., Fan C., Pei H.* Optochemical Control of DNA-Switching Circuits for Logic and Probabilistic Computation. Angew. Chem. Int. Ed. 2021, 60(7), 3397-3401.⭐封面文章

32. Lai W., Xiao M., Yang H., Li L., Fan C., Pei H.* Circularized blocker-displacement amplification for multiplex detection of rare DNA variants. Chem. Commun. 2020, 56(82), 12331-12334.⭐封面文章

31. Yang H., Xiao M., Lai W., Wan Y., Li L., Pei H.* Stochastic DNA Dual-Walkers for Ultrafast Colorimetric Bacteria Detection. Anal. Chem. 2020, 92(7), 4990-4995.
30. Cao M., Sun Y., Xiao M., Li L., Liu X., Jin H., Pei H.* Multivalent Aptamer-modified DNA Origami as Drug Delivery System for Targeted Cancer Therapy. Chem. Res. Chinese Universities. 2020, 36(2), 254-260.
29. Xiao M., Lai W., Wang F., Li L., Fan C., Pei H.* Programming Drug Delivery Kinetics for Active Burst Release with DNA Toehold Switches. J. Am. Chem. Soc. 2019, 141(51), 20354-20364.
28. Xiao M., Wang X., Li L., Pei H.* Stochastic RNA Walkers for Intracellular MicroRNA Imaging. Anal. Chem. 2019, 91(17), 11253-11258.
27. Xiao M.+, Zou K.+, Li L., Wang L., Tian Y., Fan C., Pei H.* Inside Back Cover: Stochastic DNA Walkers in Droplets for Super‐Multiplexed Bacterial Phenotype Detection. Angew. Chem. Int. Ed. 2019, 58(43), 15553-15553. ⭐当期封底内页
26. Yu H., Man T., Ji W., Shi L., Wu C., Pei H.*, Zhang C.* Controllable self-assembly of parallel gold nanorod clusters by DNA origami. Chinese Chemical Letters. 2019, 30(1), 175-178.
25. Shen J.+, Liang L.+, Xiao M., Xie X., Wang F., Li Q., Ge Z., Li J., Shi J., Wang L., Li L., Pei H.*, Fan C.* Fractal Nanoplasmonic Labels for Supermultiplex Imaging in Single Cells. J. Am. Chem. Soc. 2019, 141(30), 11938-11946.
24. Su Y., Li D., Liu B., Xiao M., Wang F., Li L., Zhang X., Pei H.* Rational Design of Framework Nucleic Acids for Bioanalytical Applications. Chempluschem. 2019, 84(5), 512-523.
23. Tang Q.+, Plank T.N.+, Zhu T., Yu H., Ge Z., Li Q., Li L., Davis J.T.*, Pei H.* Self-Assembly of Metallo-Nucleoside Hydrogels for Injectable Materials That Promote Wound Closure. ACS Appl. Mater. Interfaces. 2019, 11(22), 19743-19750.
22. Xiao M., Lai W., Man T., Chang B., Li L., Chandrasekaran A.R.*, Pei H.* Rationally Engineered Nucleic Acid Architectures for Biosensing Applications. Chem. Rev. 2019, 119(22), 11631-11717.
21. Man T., Ji W., Liu X., Zhang C.*, Li L., Pei H.*, Fan C.* Chiral Metamolecules with Active Plasmonic Transition. ACS Nano. 2019, 13(4), 4826-4833.
20. Pei H.*, Sha R., Wang X., Zheng M., Fan C.*, Canary J.W., Seeman N.C.* Organizing End-Site-Specific SWCNTs in Specific Loci Using DNA. J. Am. Chem. Soc. 2019, 141(30), 11923-11928.
19. Lai W., Xiong X., Wang F., Li Q., Li L., Fan C., Pei H.* Nonlinear Regulation of Enzyme-Free DNA Circuitry with Ultrasensitive Switches. ACS Synth. Biol. 2019, 8(9), 2106-2112.
18. Huang P.+, Wang J.+, Jiao L., Gu D., Jiang S., Li M., Lv W., Chen H.*, Pei H.* A time-frozen technique in microchannel used for the thermodynamic studies of DNA origami. Biosens. Bioelectron. 2019, 131, 224-231.
17. Deng S.+, Yan J.+, Wang F., Su Y., Zhang X., Li Q., Liu G., Fan C., Pei H.*, Wan Y.* In situ terminus-regulated DNA hydrogelation for ultrasensitive on-chip microRNA assay. Biosens. Bioelectron. 2019, 137, 263-270.
16. Lai W.+, Ren L.+, Tang Q., Qu X., Li J., Wang L., Li L., Fan C., Pei H.* Programming Chemical Reaction Networks Using Intramolecular Conformational Motions of DNA. ACS Nano. 2018, 12(7), 7093-7099.
15. Zhong R., Tang Q., Wang S., Zhang H., Zhang F.*, Xiao M., Man T., Qu X., Li L., Zhang W., Pei H.* Self-Assembly of Enzyme-Like Nanofibrous G-Molecular Hydrogel for Printed Flexible Electrochemical Sensors. Adv. Mater. 2018, 30(12), e1706887.
14. Chen L.+, Chao J.+, Qu X.*, Zhang H., Zhu D., Su S., Aldalbahi A., Wang L.*, Pei H.* Probing Cellular Molecules with PolyA-Based Engineered Aptamer Nanobeacon. ACS Appl. Mater. Interfaces. 2017, 9(9), 8014-8020.
13. Qi L., Xiao M., Wang X., Wang C., Wang L., Song S., Qu X., Li L., Shi J., Pei H.* DNA-Encoded Raman-Active Anisotropic Nanoparticles for microRNA Detection. Anal. Chem.2017, 89(18), 9850-9856.
12. Qu X.+, Wang S.+, Ge Z.+, Wang J., Yao G., Li J., Zuo X., Shi J., Song S., Wang L., Li L., Pei H.*, Fan C.* Programming Cell Adhesion for On-Chip Sequential Boolean Logic Functions. J. Am. Chem. Soc. 2017, 139(30), 10176-10179.
11. Qu X.+, Yang F.+, Chen H.*, Li J., Zhang H., Zhang G., Li L., Wang L., Song S., Tian Y., Pei H.* Bubble-Mediated Ultrasensitive Multiplex Detection of Metal Ions in Three-Dimensional DNA Nanostructure-Encoded Microchannels. ACS Appl. Mater. Interfaces. 2017, 9(19), 16026-16034.
10. Qu X., Zhang H., Chen H.*, Aldalbahi A., Li L., Tian Y., Weitz D.A., Pei H.* Convection-Driven Pull-Down Assays in Nanoliter Droplets Using Scaffolded Aptamers. Anal. Chem. 2017, 89(6), 3468-3473.
9. Qu X.+, Zhu D.+, Yao G.+, Su S., Chao J., Liu H., Zuo X., Wang L., Shi J., Wang L., Huang W., Pei H.*, Fan C.* An Exonuclease III-Powered, On-Particle Stochastic DNA Walker. Angew. Chem. Int. Ed. 2017, 56(7), 1855-1858.
8. Zhu D., Pei H., Yao G., Wang L., Su S., Chao J., Wang L., Aldalbahi A., Song S., Shi J., Hu J., Fan C., Zuo X.* A Surface-Confined Proton-Driven DNA Pump Using a Dynamic 3D DNA Scaffold. Adv. Mater. 2016, 28(32), 6860-6865.
7. Zhu D., Song P., Shen J., Su S., Chao J., Aldalbahi A., Zhou Z., Song S., Fan C., Zuo X., Tian Y., Wang L., Pei H.* PolyA-Mediated DNA Assembly on Gold Nanoparticles for Thermodynamically Favorable and Rapid Hybridization Analysis. Anal. Chem. 2016, 88(9), 4949-4954.
6. Zhu D.+, Pei H.+, Chao J., Su S., Aldalbahi A., Rahaman M., Wang L., Wang L., Huang W., Fan C., and Zuo X.* Poly-adenine-based programmable engineering of gold nanoparticles for highly regulated spherical DNAzymes. Nanoscale. 2015, 7(44), 18671-18676.
5. Pei H., Zuo X., Zhu D., Huang Q., Fan C. * (2014) Functional DNA nanostructures for theranostic applications. Acc. Chem. Res. 47, 550–559. (邀请综述)
4. Pei, H., Li, F., Wan Y., Wei M., Liu H.*, Su Y., Chen N., Huang Q., Fan C.* (2012) Designed Diblock Oligonucleotide for the Synthesis of Spatially Isolated and Highly Hybridizable Functionalization of DNA–Gold Nanoparticle Nanoconjugates. J. Am. Chem. Soc.134, 11876-11879.
☆Highlighted by the editors in JACS spotlights with “DNA−Gold Nanoparticle Combo gets Straight A’s”(该工作被JACS Spotlights 作为亮点文章报道)
3. Pei, H., Liang, L., Yao G., Li J., Huang Q., Fan C.* (2012) Reconfigurable three-dimensional DNA nanostructures for construction of intracellular logistic sensors. Angew. Chem. Int. Ed. 51, 9020-9024.
☆Chosen by the editors as Back Inside Cover; Highlight in Nature China (该工作被选为当期封底,并被Nature China报道)
2. Pei, H., Li J., Lv M., Wang J., Gao J., Lv J, Li Y., Huang Q., Hu J.*, Fan C.* (2012) A graphene-based sensor array for high-precision and adaptive target identification with ensemble aptamers. J. Am. Chem. Soc.134, 13843-13849.
1. Pei, H., Lu, N., Wen, Y., Song, S., Liu, Y., Yan, H. * and Fan, C.* (2010) A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 22, 4754-4758.
☆This paper was chosen by editors as cover story. (该工作以封面文章报道)
